RFC-2: Zarr v3#
Adopt the version 3 of Zarr for OME-Zarr.
Status#
This RFC is currently in SPEC state (S1).
Role |
Name |
GitHub Handle |
Institution |
Date |
Status |
|---|---|---|---|---|---|
Author |
Norman Rzepka |
2024-02-14 |
|||
Endorser |
Davis Bennett |
2024-02-14 |
Endorse |
||
Endorser |
Kevin Yamauchi |
ETH Zürich |
2024-02-16 |
||
Endorser |
John Bogivic |
HHMI Janelia Research Campus |
2024-02-16 |
||
Endorser |
Matthew Hartley |
EMBL-EBI |
2024-02-16 |
||
Endorser |
Christian Tischer |
EMBL |
2024-02-16 |
||
Endorser |
Joel Lüthi |
BioVisionCenter, University of Zurich |
2024-02-16 |
||
Endorser |
Constantin Pape |
University Göttingen |
2024-02-18 |
||
Endorser |
Will Moore |
OME, University of Dundee |
2024-02-19 |
||
Endorser |
Juan Nunez-Iglesias |
Biomedicine Discovery Institute, Monash University |
2024-02-20 |
||
Endorser |
Eric Perlman |
2024-02-22 |
|||
Endorser |
Ziwen Liu |
Chan Zuckerberg Biohub |
2024-03-12 |
||
Endorser |
Lachlan Deakin |
Australian National University |
2024-03-14 |
||
Reviewer |
Melissa Linkert, Sébastien Besson, Chris Allan, Jason Swedlow |
glencoesoftware |
Glencoe Software |
2024-05-23 |
|
Reviewer |
Yaroslav O. Halchenko |
yarikoptic |
Dartmouth College, DANDI Project |
2024-06-10 |
|
Reviewer |
Jeremy Maitin-Shepard |
jbms |
2024-04-30 |
||
Reviewer |
Melissa Linkert, Sébastien Besson, Chris Allan, Jason Swedlow |
glencoesoftware |
Glencoe Software |
2024-08-05 |
|
Reviewer |
Jeremy Maitin-Shepard |
jbms |
2024-09-11 |
||
Reviewer |
Yaroslav O. Halchenko |
yarikoptic |
Dartmouth College, DANDI Project |
2024-09-11 |
Overview#
This RFC adopts Zarr v3 as the new underlying format of OME-Zarr.
Background#
OME-Zarr uses the Zarr format as underlying data format. Zarr is not only used for bioimaging data but also in several other communities, such as astronomy, geo, earth and climate sciences. There is a governance structure around Zarr that structures the evolution of the format.
In summer 2023, version 3 of the Zarr specification has been accepted by the Zarr implementation and steering councils through the ZEP process. A major motivation for the new version is the introduction of extension hooks.
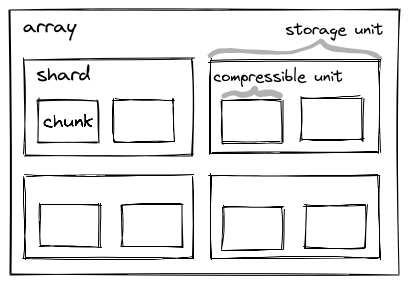
One of these extensions is the sharding codec that has also been accepted by the Zarr councils. Sharding provides a mechanism to store multiple chunks within one file/object. This can greatly reduce the number of files/objects that are required for large Zarr arrays, while preserving fast access to individual chunks and parallel writing capabilities. Sharding solves some pain points that are also greatly felt by users in the OME-Zarr community. Adopting Zarr v3 in OME-Zarr is a precondition for using sharding.
Library support for Zarr v3 is already available for several languages:
Visualization tools with integrated Zarr v3 implementations are also available:
Support for other languages is under active development.
Libraries will likely prioritize support for v3 over previous versions in the near future. OME-Zarr should therefore adopt the new version for future-proofing.
Other notable changes in Zarr v3#
There are a few notable changes that Zarr v3 brings for OME-Zarr:
Array and group metadata including attributes are now stored in
zarr.jsonfiles instead of.zarray,.zgroupand.zattrs. The attributes are now represented in anattributeskey within thezarr.jsonfiles.Arrays specify a
chunk_key_encodingthat controls under what naming scheme chunks are stored. This is similar to the previousdimension_separatorattribute. As part of this proposal, OME-Zarr will support all valid chunk key encodings instead of mandating a/dimension separator.There is a new codec pipeline concept that unifies filters and compression codecs as well as array-to-byte serialization including endianness and index ordering configuration. OME-Zarr will support all codecs in the specification. In the future there will likely be additional codecs including image-specific codecs that OME-Zarr would automatically adopt. This is the current list of available codecs:
blosc for compression
gzip for compression
transpose for transposing the data before serialization, e.g. to support C and F orders
bytes for serializing arrays to byte streams with configurable endianness
crc32c for decorating chunks with a checksum
sharding_indexed for storing sharded arrays (see below).
The Zarr specification does not prescribe the support stores for Zarr hierarchies. HTTP(S), File system, S3, GCS, and Zip files are commonly used stores.
Proposal#
This RFC proposes to adopt version 3 of the Zarr format for OME-Zarr. Images that use the new version of OME-Zarr metadata MUST NOT use Zarr version 2 any more.
With this proposal all features of the Zarr specification are allowed in OME-Zarr. In the future, the OME-Zarr community MAY decide to restrict the allowed feature set.
The motivation for making this hard cut is to reduce the burden of complexity for implementations. Currently, many Zarr library implementations support both versions. However, in the future they might deprecate support for version 2 or deprioritize it in terms of features and performance. Additionally, there are OME-Zarr implementations that have their own integrated Zarr stack. With this hard cut, implementations that only support OME-Zarr versions ≥ 0.5 will not need to implement Zarr version 2 as well.
From an OME-Zarr user perspective, the hard cut also makes things simpler: < 0.5 => Zarr version 2 and ≥ 0.5 => Zarr version 3. If users wish to upgrade their data from one OME-Zarr version to another, migration tools will be available (prototype here). Migration is a fairly computationally cheap operation, because only json files are touched.
Due to the existence of large quantities of images in OME-Zarr 0.4, it is RECOMMENDED that implementations continue to support OME-Zarr 0.4 with the underlying Zarr v2.
OME-Zarr images MUST be consistent in their OME-Zarr and Zarr version. With this constraint, implementations only need to detect the version of a provided URL or file path once and can assume that all multiscale levels, wells, series images etc. use the same version.
While technically possible, OME-Zarr 0.5 (with Zarr v3) and OME-Zarr 0.4 (with Zarr v2) metadata could exist side-by-side in a Zarr hierarchy, it is NOT RECOMMENDED. This may be useful for short periods of time (i.e. during migrations from 0.4 to 0.5), but should not be used longer term. Multiple metadata versions can lead to conflicts, which may be hard to resolve by implementations. If implementations encounter 0.4 and 0.5 metadata side-by-side, 0.5 SHOULD be treated preferentially.
Changes to the OME-Zarr metadata#
While the adoption of Zarr v3 does not strictly require changes to the OME-Zarr metadata, this proposal contains changes to align with community conventions and ease implementation:
OME-Zarr metadata will be stored under a dedicated
omekey in the Zarr array or group attributes.The version information will be moved from the
multiscale,plate,welletc. sections into the newomesection.The
dimension_namesattribute in the Zarr metadata must match the axes names in the OME-Zarr metadata.
Finally, this proposal changes the title of the OME-Zarr specification document to “OME-Zarr specification”.
Requirements#
The key words “MUST”, “MUST NOT”, “REQUIRED”, “SHALL”, “SHALL NOT”, “SHOULD”, “SHOULD NOT”, “RECOMMENDED”, “MAY”, and “OPTIONAL” in this document are to be interpreted as described in IETF RFC 2119
Stakeholders#
Preliminary work of this RFC has been discussed in:
this image.sc post
this image.sc post
this image.sc post
this pull request
several Zarr community calls
several recent OME-NGFF community calls.
Implementation#
OME-Zarr implementations can rely on existing Zarr libraries to implement the adoption of Zarr v3. See Background for a list of v3-capable Zarr libraries.
Support for the OME-Zarr 0.5 metadata is under development in ome-zarr-py and other implementations.
ngff-zarr supports creating OME-Zarr 0.5 from Python via Zarr-Python or Tensorstore, converting OME-Zarr version 0.4 to 0.5 and 0.5 to 0.4, validating OME-Zarr 0.5 metadata, and converting other file formats to OME-Zarr 0.5.
Drawbacks, risks, alternatives, and unknowns#
While it is clear that Zarr v3 will become the predominant version of the specification moving forward, current library support for v3 is still under active development.
An alternative to this proposal would be to add Zarr v3 support to OME-Zarr 0.4 without changes to the OME-Zarr Metadata.
The contents of the .zattrs would simply move to the attributes within the zarr.json.
There would need to be some transparency for users to know what Zarr versions are supported by an implementation.
Additionally, there would be no opportunity to introduce an ome namespace in the attributes that is useful for composability.
Performance#
The adoption of Zarr v3 will not necessarily have an impact on performance. The performance is determined by a wide range of parameters, many of which are specific to implementations.
Using sharding can have a profound impact on the number of files/objects that large images consume. On some storage backends using less files/objects can be beneficial for the performance of various operations. In particular, the chunk sizes can be made small to facilitate interactive visualization without incurring the overhead of too many files/objects.
Backwards Compatibility#
The metadata of Zarr v3 arrays is not backwards compatible with that of Zarr v2 arrays.
Implementations of OME-Zarr MUST specify the version(s) of the OME-Zarr specification that they support.
It is RECOMMENDED that implementations of OME-Zarr that support both v2 and v3-based OME-Zarr versions auto-detect the underlying Zarr version.
While the metadata of Zarr v3 is not backwards compatible, the chunk data is largely backwards compatible, only depending on compressor configuration. There are scripts available to migrate Zarr v2 metadata to Zarr v3.
Abandoned Ideas#
Previous versions of this proposal contained changes to referencing labels in the OME-Zarr metadata.
This has been delayed to future RFCs.
Previous versions of this proposal have used a versioned namespace, e.g. https://ngff.openmicroscopy.org/0.5, in the Zarr attributes instead of a simple ome namespace with dedicated version attribute.
This has been abandoned because it makes discovery of versions more difficult.
Additionally, handling of multiple versions may be ill-defined.
Examples#
File hierarchy of one multi-scale OME-Zarr image 456.zarr:
456.zarr
│
├── zarr.json
├── 1
│ ├── zarr.json
│ └─ c
│ ├─ 0
│ | ├─ 0
│ | | ├─ 0
│ | | | ├─ 0
│ | | | └─ ...
│ | | └─ ...
│ | └─ ...
│ └─ ...
│ ...
└── n
456.zarr/zarr.json:
{
"zarr_format": 3,
"node_type": "group",
"attributes": {
"ome": {
"version": "0.5",
"multiscales": [
{
"axes": [
{
"name": "c",
"type": "channel"
},
{
"name": "x",
"type": "space",
"unit": "nanometer"
},
{
"name": "y",
"type": "space",
"unit": "nanometer"
},
{
"name": "z",
"type": "space",
"unit": "nanometer"
}
],
"datasets": [
{
"path": "1",
"coordinateTransformations": [
{
"type": "scale",
"scale": [1.0, 11.24, 11.24, 28.0]
}
]
},
{
"path": "2-2-1",
"coordinateTransformations": [
{
"type": "scale",
"scale": [1.0, 22.48, 22.48, 28.0]
}
]
},
{
"path": "4-4-1",
"coordinateTransformations": [
{
"type": "scale",
"scale": [1.0, 44.96, 44.96, 28.0]
}
]
},
{
"path": "8-8-2",
"coordinateTransformations": [
{
"type": "scale",
"scale": [1.0, 89.92, 89.92, 56.0]
}
]
},
{
"path": "16-16-4",
"coordinateTransformations": [
{
"type": "scale",
"scale": [1.0, 179.84, 179.84, 112.0]
}
]
}
]
}
]
}
}
}
456.zarr/1/zarr.json:
{
"zarr_format": 3,
"node_type": "array",
"shape": [1, 4096, 4096, 1536],
"data_type": "uint8",
"chunk_grid": {
"configuration": { "chunk_shape": [1, 1024, 1024, 1024] },
"name": "regular"
},
"chunk_key_encoding": {
"configuration": { "separator": "/" },
"name": "default"
},
"fill_value": 0,
"codecs": [
{
"configuration": {
"chunk_shape": [1, 32, 32, 32],
"codecs": [
{ "name": "transpose", "configuration": { "order": [3, 2, 1, 0] } },
{ "name": "bytes" },
{
"name": "blosc",
"configuration": {
"typesize": 1,
"cname": "zstd",
"clevel": 5,
"shuffle": "noshuffle",
"blocksize": 0
}
}
],
"index_codecs": [{ "name": "bytes" }, { "name": "crc32c" }],
"index_location": "end"
},
"name": "sharding_indexed"
}
],
"attributes": {},
"dimension_names": ["c", "x", "y", "z"]
}