Spectral clustering#
You can use Laplace spectra to cluster data. The idea is that the eigenvalues of the mesh’s Laplace spectrum can be embedded in a lower-dimensional space.
Data source for this notebooks: odedstein/meshes
import os
import glob
import napari_shape_odyssey as nso
import vedo
import numpy as np
import tqdm
from sklearn.manifold import TSNE
from sklearn.cluster import DBSCAN
import napari
import pandas as pd
import matplotlib.pyplot as plt
!git clone https://github.com/odedstein/meshes.git
Cloning into 'meshes'...
Updating files: 17% (17/96)
Updating files: 18% (18/96)
Updating files: 19% (19/96)
Updating files: 20% (20/96)
Updating files: 21% (21/96)
Updating files: 22% (22/96)
Updating files: 23% (23/96)
Updating files: 25% (24/96)
Updating files: 26% (25/96)
Updating files: 27% (26/96)
Updating files: 28% (27/96)
Updating files: 29% (28/96)
Updating files: 30% (29/96)
Updating files: 31% (30/96)
Updating files: 32% (31/96)
Updating files: 33% (32/96)
Updating files: 34% (33/96)
Updating files: 35% (34/96)
Updating files: 36% (35/96)
Updating files: 37% (36/96)
Updating files: 38% (37/96)
Updating files: 39% (38/96)
Updating files: 40% (39/96)
Updating files: 41% (40/96)
Updating files: 42% (41/96)
Updating files: 43% (42/96)
Updating files: 44% (43/96)
Updating files: 45% (44/96)
Updating files: 46% (45/96)
Updating files: 47% (46/96)
Updating files: 48% (47/96)
Updating files: 50% (48/96)
Updating files: 51% (49/96)
Updating files: 52% (50/96)
Updating files: 53% (51/96)
Updating files: 54% (52/96)
Updating files: 55% (53/96)
Updating files: 56% (54/96)
Updating files: 57% (55/96)
Updating files: 58% (56/96)
Updating files: 59% (57/96)
Updating files: 60% (58/96)
Updating files: 61% (59/96)
Updating files: 62% (60/96)
Updating files: 63% (61/96)
Updating files: 64% (62/96)
Updating files: 65% (63/96)
Updating files: 66% (64/96)
Updating files: 67% (65/96)
Updating files: 68% (66/96)
Updating files: 69% (67/96)
Updating files: 70% (68/96)
Updating files: 71% (69/96)
Updating files: 72% (70/96)
Updating files: 73% (71/96)
Updating files: 75% (72/96)
Updating files: 76% (73/96)
Updating files: 77% (74/96)
Updating files: 78% (75/96)
Updating files: 79% (76/96)
Updating files: 80% (77/96)
Updating files: 81% (78/96)
Updating files: 82% (79/96)
Updating files: 83% (80/96)
Updating files: 84% (81/96)
Updating files: 85% (82/96)
Updating files: 86% (83/96)
Updating files: 87% (84/96)
Updating files: 88% (85/96)
Updating files: 89% (86/96)
Updating files: 90% (87/96)
Updating files: 91% (88/96)
Updating files: 92% (89/96)
Updating files: 93% (90/96)
Updating files: 94% (91/96)
Updating files: 95% (92/96)
Updating files: 96% (93/96)
Updating files: 97% (94/96)
Updating files: 98% (95/96)
Updating files: 100% (96/96)
Updating files: 100% (96/96), done.
mesh_files = glob.glob(os.path.join('./meshes/**/*.obj'))
viewer = napari.Viewer()
Calculate spectra for every mesh#
eigenvalues = []
eigenvectors = []
meshes = []
for file in tqdm.tqdm(mesh_files):
# load and preprocess meshes
mesh = vedo.load(file).triangulate().clean().decimate(n=5000)
bounding_box_volume = np.prod(mesh.bounds()[1] - mesh.bounds()[0])
mesh.scale(1 / bounding_box_volume)
mesh_tuple = (mesh.points(), np.asarray(mesh.faces()))
meshes.append(mesh_tuple)
viewer.add_surface(mesh_tuple, name = os.path.basename(file))
_eigenvectors, _eigenvalues = nso.spectral.shape_fingerprint(mesh_tuple, order = 20)
eigenvalues.append(_eigenvalues)
eigenvectors.append(_eigenvectors)
# stack eigenvalues into dataframe
eigenvalues_stack = np.stack(eigenvalues)
df = pd.DataFrame(eigenvalues_stack, columns = [f'lambda_{i}' for i in range(eigenvalues_stack.shape[1])])
df.head()
| lambda_0 | lambda_1 | lambda_2 | lambda_3 | lambda_4 | lambda_5 | lambda_6 | lambda_7 | lambda_8 | lambda_9 | lambda_10 | lambda_11 | lambda_12 | lambda_13 | lambda_14 | lambda_15 | lambda_16 | lambda_17 | lambda_18 | lambda_19 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -1.953646e-14 | 3.497411 | 4.534560 | 5.684497 | 9.985113 | 14.893090 | 22.529184 | 29.669639 | 30.541655 | 37.723114 | 41.284098 | 52.551030 | 58.486390 | 63.681724 | 64.850003 | 71.539644 | 78.696079 | 81.764765 | 95.658377 | 102.104163 |
| 1 | 1.382054e-14 | 3.497411 | 4.534560 | 5.684497 | 9.985113 | 14.893090 | 22.529184 | 29.669640 | 30.541655 | 37.723113 | 41.284097 | 52.551029 | 58.486391 | 63.681721 | 64.850001 | 71.539641 | 78.696077 | 81.764764 | 95.658375 | 102.104161 |
| 2 | -2.596881e-15 | 0.683063 | 2.141403 | 4.065282 | 4.768848 | 4.828398 | 7.341017 | 7.398343 | 7.880799 | 10.123669 | 10.174560 | 11.785636 | 13.405222 | 14.186425 | 15.805213 | 16.056310 | 16.568611 | 18.689970 | 19.308144 | 21.437427 |
| 3 | -3.480202e-14 | 7.361894 | 14.320784 | 29.855033 | 33.864331 | 38.218302 | 51.627914 | 55.100373 | 63.052792 | 71.963617 | 75.823578 | 91.888147 | 95.434760 | 99.987659 | 114.406247 | 123.437142 | 127.149471 | 140.136072 | 142.793974 | 153.672580 |
| 4 | 8.763823e-15 | 5.616643 | 7.917237 | 8.444330 | 16.360097 | 19.160159 | 20.802747 | 22.530132 | 30.035678 | 35.645911 | 37.769361 | 40.187326 | 41.239513 | 45.143890 | 48.583533 | 55.172578 | 60.318931 | 60.867925 | 64.006032 | 70.048105 |
Normalize spectra#
# Fit a linear curve to every row of the dataframe and divide every value in the row by the slope of the curve
slope = []
intercept = []
for i in range(0, len(df)):
x = np.arange(0, len(df.columns))
y = df.iloc[i].values
m, b = np.polyfit(x, y, 1)
slope.append(m)
intercept.append(b)
df = df.div(slope, axis=0)
Dimensionality reduction#
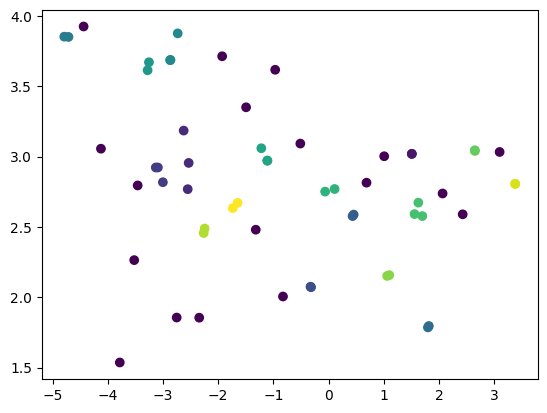
embedder = TSNE(n_components=2)
X_embedded = embedder.fit_transform(df)
clusterer = DBSCAN(eps=0.25, min_samples=2)
clusterer.fit(X_embedded)
for idx, layer in enumerate(viewer.layers):
mesh_tuple = list(layer.data)
mesh_tuple[-1] *= clusterer.labels_[idx]
viewer.layers[idx].data = tuple(mesh_tuple)
viewer.layers[idx].colormap = 'tab10'
viewer.layers[idx].contrast_limits = [-1, np.max(clusterer.labels_)]
plt.scatter(X_embedded[:, 0], X_embedded[:, 1], c = clusterer.labels_)
<matplotlib.collections.PathCollection at 0x155abe32bb0>
for idx in np.argwhere(clusterer.labels_==4):
viewer.add_surface(meshes[idx[0]], name = os.path.basename(mesh_files[idx[0]]))