Unwrap to disk#
import os
from skimage import io
import napari
import vedo
import numpy as np
from napari_stress import measurements, vectors
import napari_segment_blobs_and_things_with_membranes as nsbatwm
import napari_process_points_and_surfaces as nppas
import pyclesperanto_prototype as cle
from napari_shape_odyssey import unwrapping
viewer = napari.Viewer()
Invalid schema for package 'napari-stl-exporter', please run 'npe2 validate napari-stl-exporter' to check for manifest errors.
WARNING: QWindowsWindow::setGeometry: Unable to set geometry 6880x2754+1280+560 (frame: 6906x2825+1267+502) on QWidgetWindow/"_QtMainWindowClassWindow" on "\\.\DISPLAY1". Resulting geometry: 3844x2119+1280+560 (frame: 3870x2190+1267+502) margins: 13, 58, 13, 13 minimum size: 385x499 MINMAXINFO maxSize=0,0 maxpos=0,0 mintrack=796,1069 maxtrack=0,0)
root = r'C:\Users\johamuel\Desktop\test_surface_nuclei_unwrapping'
filename = r'Strausberg_Tribolium_LA-GFP_tailpole_run-C0opticsprefused-301-310.tif'
image = io.imread(os.path.join(root, filename))[:, :-2]
viewer.add_image(image[0], name='data', colormap='gray')
<Image layer 'data' at 0x22d8c70cb20>
image.shape
(10, 109, 1024, 1024)
image_binary = cle.threshold_otsu(image[0])
image_eroded = cle.opening_sphere(image_binary, radius_x=5, radius_y=5, radius_z=2)
image_closed = cle.closing_sphere(image_eroded, radius_x=5, radius_y=5, radius_z=2)
image_small_labels_removed = cle.exclude_small_labels(image_closed)
image_labelled = cle.connected_components_labeling_box(image_small_labels_removed)
# cheatcode:
image_labelled[-1] = 0
viewer.add_labels(image_labelled)
<Labels layer 'image_labelled' at 0x22d8db13eb0>
surface = nppas.label_to_surface(image_labelled, 1)
mesh = vedo.Mesh(surface).clean().smooth(50, pass_band=0.05, feature_angle=150, edge_angle=90, boundary=True).decimate(n=20000).clean()
smoothed = vedo.Mesh((mesh.points(), mesh.faces()))
smoothed.smooth(feature_angle=10, edge_angle=10, boundary=True).clean()
|
|
vedo.mesh.Mesh
|
intensities = measurements.measure_intensity_on_surface((smoothed.points(), np.asarray(smoothed.faces())), image[0],
measurement_range=-64, sampling_distance=4, center=True)
mean_intensity = np.nanmean(intensities.values, axis=1)
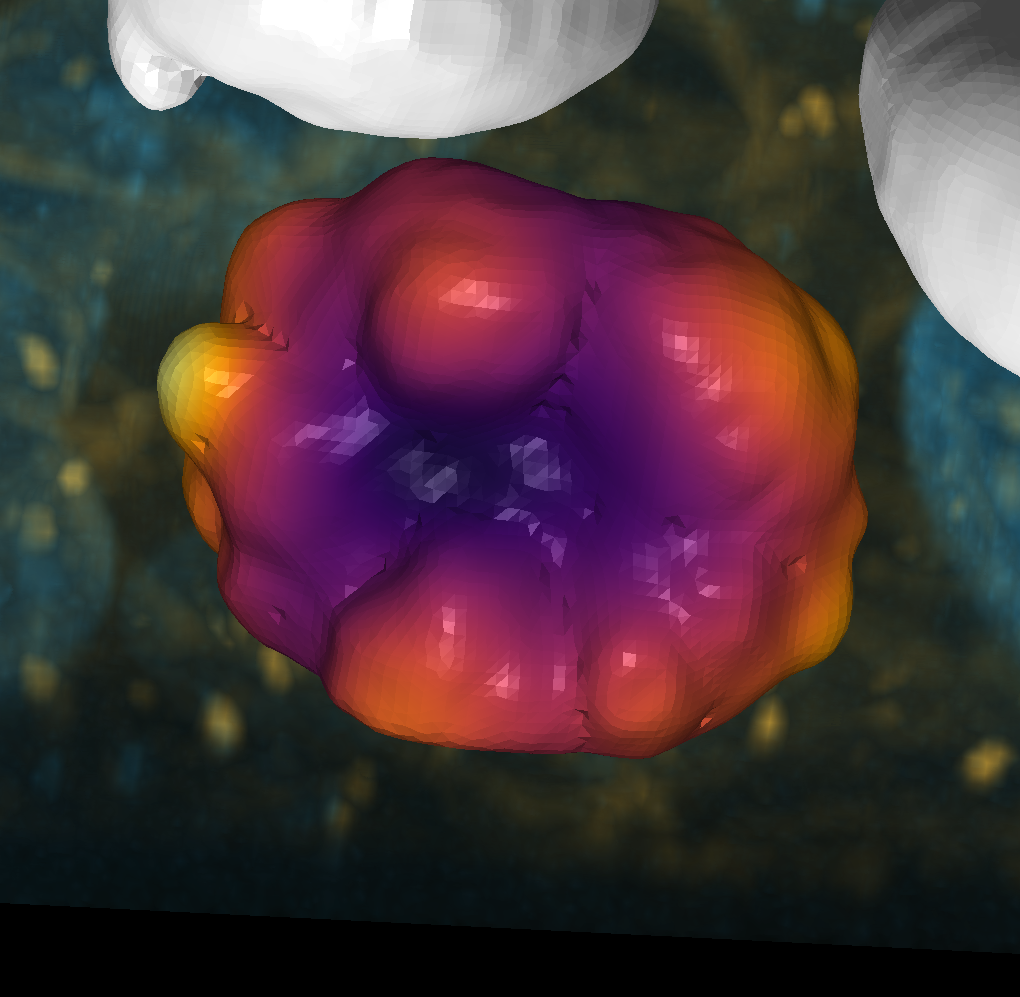
viewer.add_surface((smoothed.points(), np.asarray(smoothed.faces()), mean_intensity), colormap='inferno')
<Surface layer 'Surface' at 0x22d8c70c040>
result = unwrapping.map_surface_to_disk((smoothed.points(), np.asarray(smoothed.faces()), mean_intensity),
only_uvs=True)
new_values = np.zeros(len(result[0]))
new_values[:len(mean_intensity)] = mean_intensity
viewer.add_surface((result[0], result[1], new_values), colormap='inferno')
<Surface layer 'Surface [2]' at 0x1fa2885fd00>
result[0].shape
(20190, 3)
mean_intensity.shape
(20181,)
vedo.load(r'../../../src/napari_shape_odyssey/_unwrapping/mesh_out.obj').clean()
|
|
vedo.mesh.Mesh (...yssey/_unwrapping/mesh_out.obj)
|
vedo.load(r'../../../src/napari_shape_odyssey/_unwrapping/mesh_in.obj').clean()
|
|
vedo.mesh.Mesh (...dyssey/_unwrapping/mesh_in.obj)
|
test_out = vedo.load(os.path.join(r'C:\Users\johamuel\Downloads\windows-v1.6', 'test_out.obj'))
test = vedo.load(os.path.join(r'C:\Users\johamuel\Downloads\windows-v1.6', 'test.obj'))
test.npoints
121567
test_out.npoints
122517